| Submitted by: | Maria del Sagrario Rossano Tapia |
| Department: | Chemistry |
| Faculty: | Science |
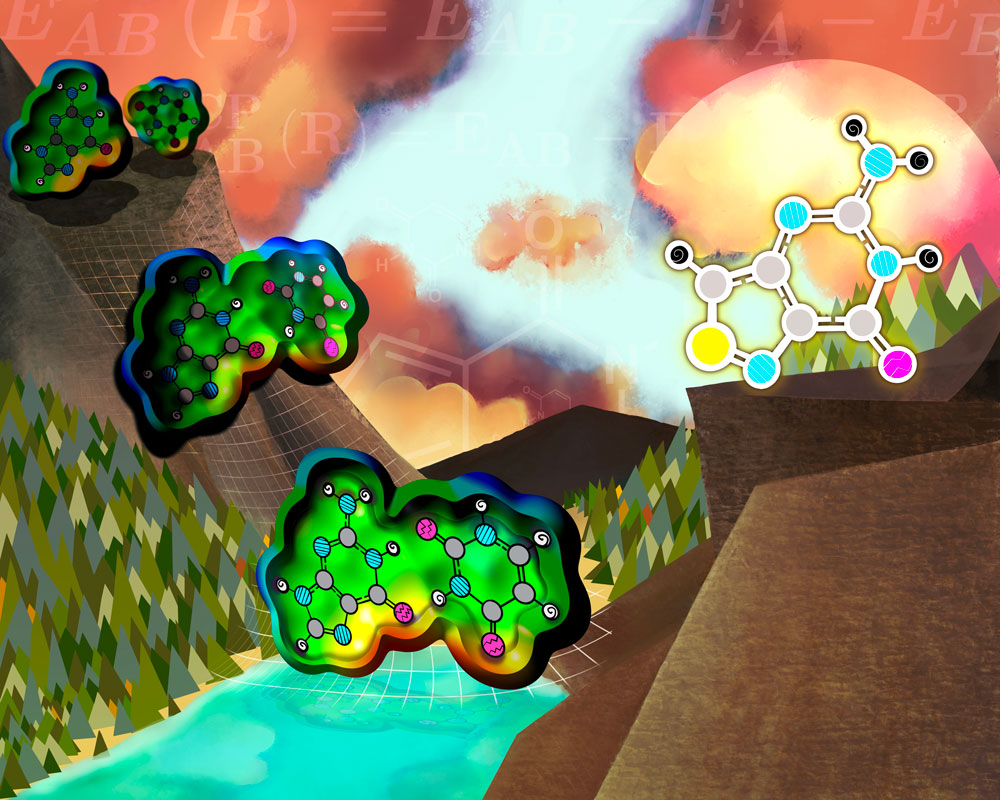
Among clinical imaging techniques it is common to monitor cellular biochemical processes using “molecular lanterns” in the form of light-emitting proteins or RNA nucleobases. Naturally occurring RNA nucleobases do not emit light (fluoresce) and thus, cannot be directly used for this purpose. Alternatively, the chemical structure of natural nucleobases can be tuned to make them fluoresce, for example, by including sulfur in their composition. However, modifications of the chemical structure of natural nucleobases could disrupt the RNA biochemical functions. This image is inspired by our quantum chemistry investigation on the RNA structural disruptions caused by a sulfur-modified guanine. The computed energy “valley” leading to the binding between the natural guanine and uracil is depicted in the image. The modified guanine could be considered a “molecular lantern” candidate if its energy valley leads to a structure resembling the natural guanine one.
